
This website is free and open to all users and there is no login requirement
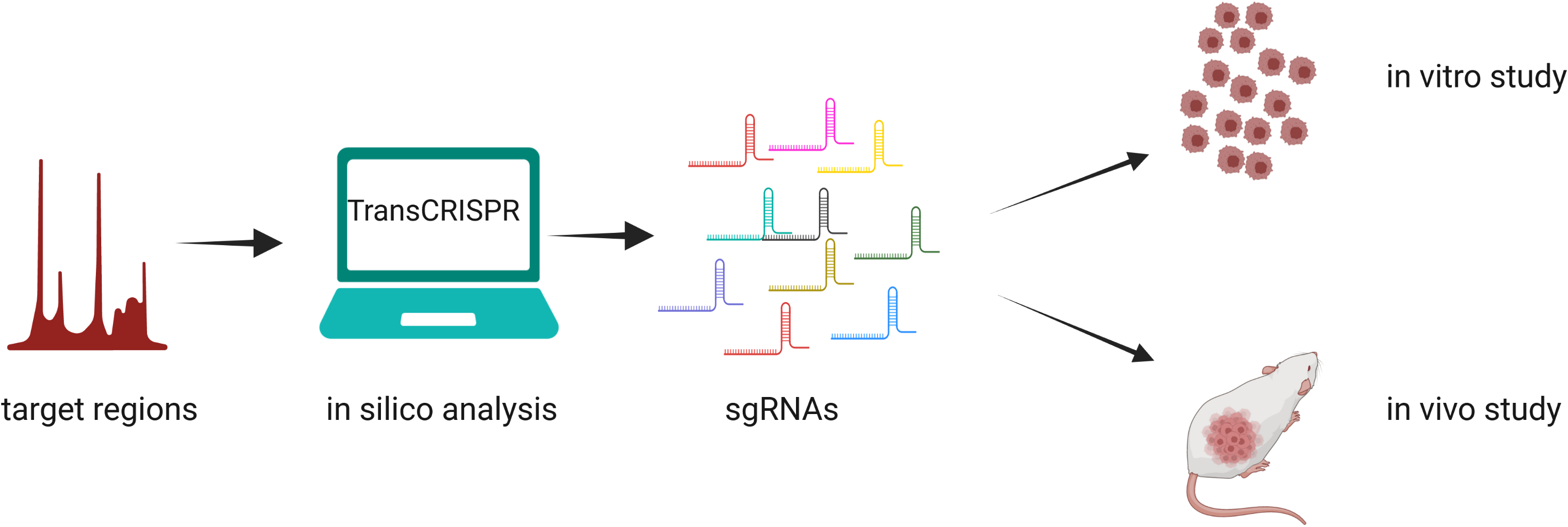
TransCRISPR is a user-friendly tool dedicated to design of sgRNAs for study of DNA sequence motifs. TransCRISPR can be applied both for single sequences as well as for large lists of genomic coordinates, enabling design of tailored CRISPR/Cas9 or dCas9 experiments. The program will search for provided DNA motifs in the query sequence and design sgRNAs targeting them. As a result, the list of identified motifs and sgRNAs along with several characteristics and statistics is generated. For more detailed program description refer to Manual or manuscript or find answers to most frequently asked questions in FAQ section.
Cas9
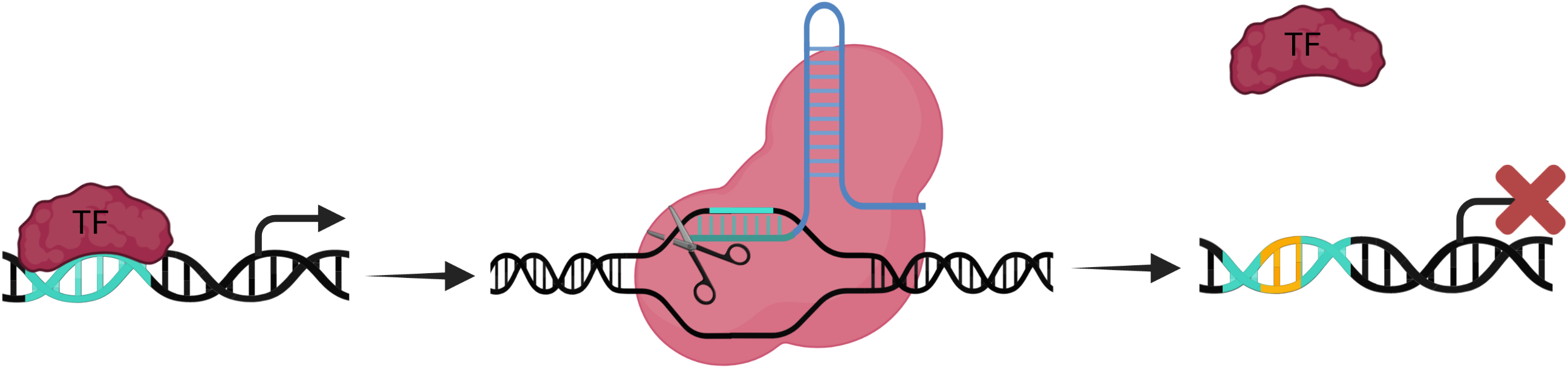
TransCRISPR will search for a given motif, e.g. transcription factor (TF) binding motif, in the provided sequence/coordinates and for the PAM (protospacer adjacent motif) within or nearby this motif. For the catalytically active Cas9 sgRNAs are designed in such way that the cut is within the TF motif (Sp Cas9 cuts DNA strands 3 nucleotides downstream of PAM). This will lead to random insertions/deletions and thus disruption of the motif.
dCas9
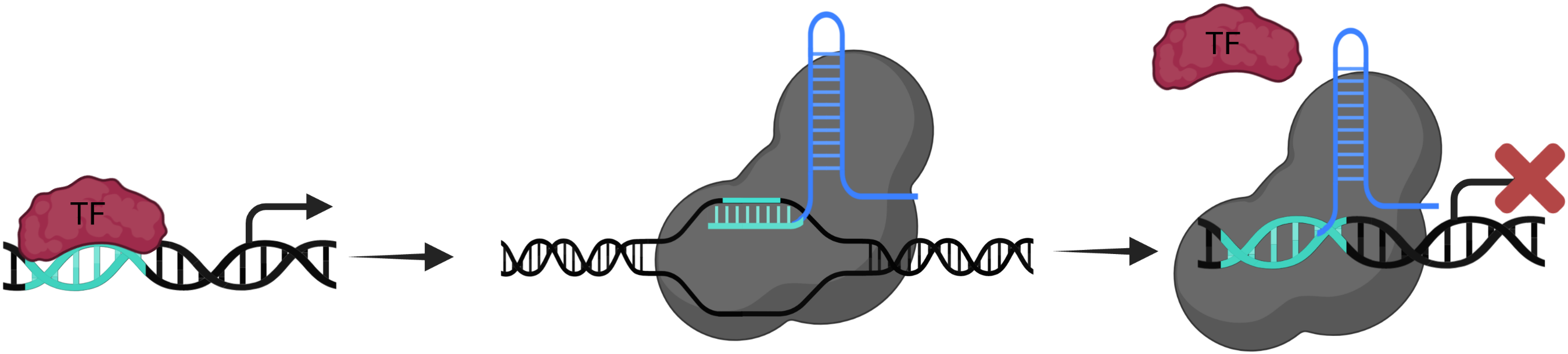
For the catalytically inactive (dead) Cas9, sgRNAs are designed in such way that they overlap at least 1 nt of the TF motif. This leads to the steric blocking of the motif, making it inaccessible for binding of the TF.
Citation
TransCRISPR-sgRNA design tool for CRISPR/Cas9 experiments targeting specific sequence motifs. Nucleic Acids Res. 2023 May 9:gkad355. doi: 10.1093/nar/gkad355


