Frequently Asked Questions (FAQ)
How does transCRISPR design sgRNAs for Cas9?
TransCRISPR will search for a given DNA motif in the provided sequence/coordinates and for the PAM (Protospacer adjacent motif) within or nearby this motif. TransCRISPR will search for PAM on both strands. Note that Sp Cas9 cuts DNA strands 3 nucleotides downstream of PAM. TransCRISPR only provides the list of sgRNAs that will lead to cut within the provided DNA motif, so that the motif can be disrupted.
How does transCRISPR design sgRNAs for dCas9?
TransCRISPR will search for a given DNA motif in the provided sequence/coordinates and for the PAM (Protospacer adjacent motif) within or nearby this motif. TransCRISPR will search for PAMon both strands. For dCas9, sgRNAs will be designed to ensure that at least 1 nt (of a 20nt guide) overlaps with the provided DNA motif, so that the motif can be sterically blocked by dCas9 protein.
How does transCRISPR design sgRNAs in the custom option?
TransCRISPR will search for a given DNA motif in the provided sequence/coordinates and for the PAM (Protospacer adjacent motif) within the range specified by the user. TransCRISPR will search for PAM on both strands. This option allows to design sgRNAs in proximity of the motif even though they may not overlap the motif.
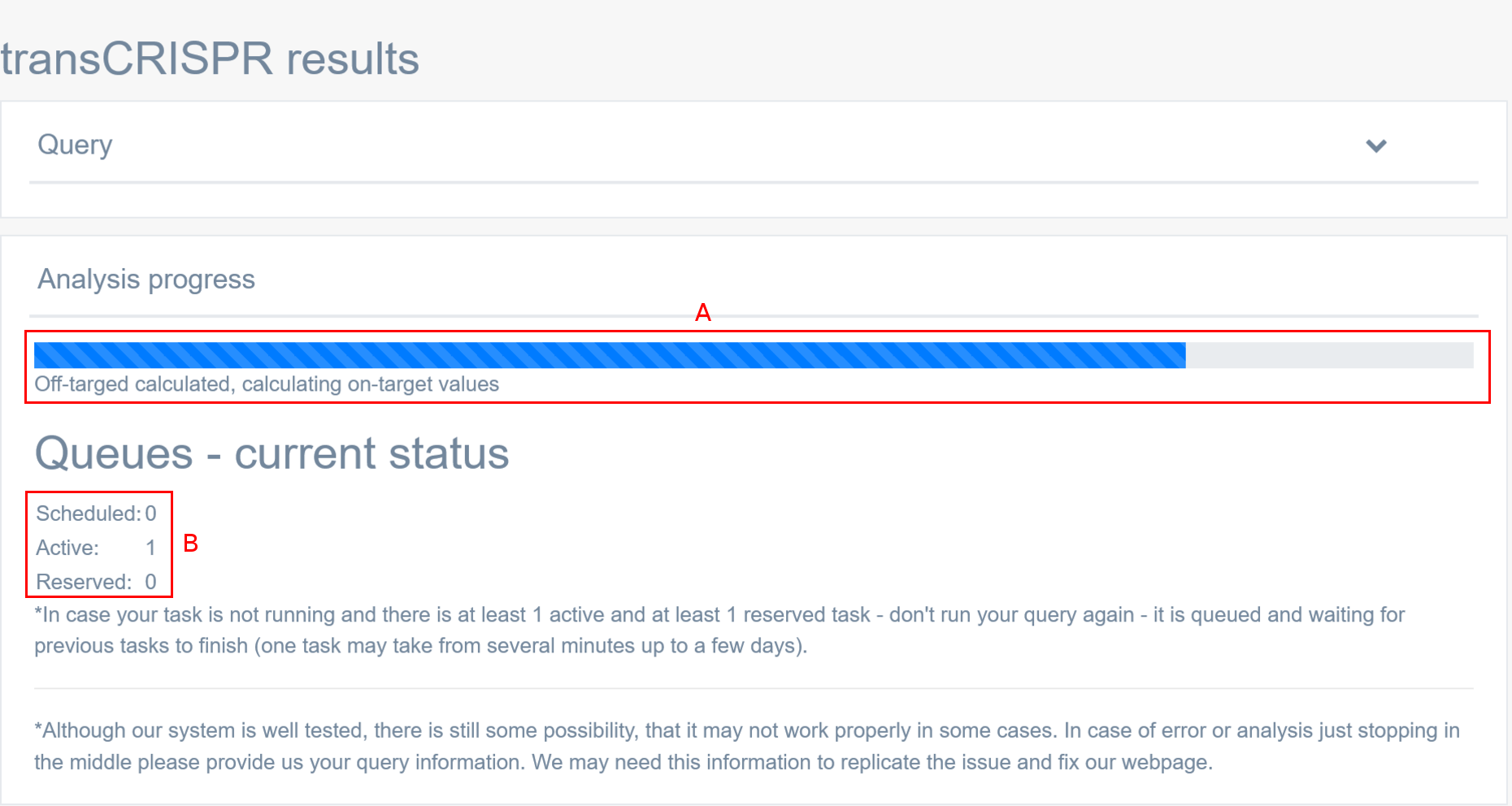
How do I know if my analysis is running?
TransCRISPR provides the real-time tracking of the analysis progress [A] and allows to see how many scheduled, active or reserved analysis are running at the same time [B]. Once the user sets up the query and clicks “Finish”, the job will be queued and will run as soon as the previous analysis is completed. Note that one task may take from several minutes up to a few days, depending on the amount of sequences to be analyzed and motifs found. User can choose to be notified by e-mail, when the analysis is finished.
How can I access the results?
For each query, a unique url address will be generated. This link can be used to access the page and check the status of the analysis as well as download the results when it is finished. The link will remain active for 7 days, since the end of the analysis.
Which PAM variants are available?
Currently transCRISPR works only for the SpCas9 NGG, NGA, NGCG PAM. We plan to include other PAM variants in the future.
Can I design guides shorter or longer than 20 nt?
No, current transCRISPR version automatically designs 20 nt guides.
Why don't I get information about the motif genomic localization and nearest genes?
These options are only available with genomic coordinates as an input. If you used DNA sequence as a text, transCRISPR will search for motifs and design sgRNAs, but information about genomic localization and closest genes will not be provided.
Can I visualize designed guides on the provided query target?
Yes, by choosing option: “Display in Genome Browser”. Note that option is available only if the user provides genomic coordinates.
What do the colors in Off/On target scores mean?
The off-target CFD score indicates the predicted off-target activity of the sgRNA. Off target score range is 0-100, the higher, the better. 100 means that no off-targets with up to 4 mismatches were found. The on-target score indicates the probability of successful cleavage for a particular guide. On target score range is 0-100, the higher, the better. Off- and on-target scores are colored as follows: 0 : ≤ 30 – red, >30 : ≤50 – yellow, >50 : ≤100 – green.
Where I can find example matrices?
- Align ace
- cluster buster
- XMS
- Meme output file motif
- minimal meme
- transfac
- four columns
- four rows
- jaspar pfm
- jaspar multiple
- jaspar sites
Is there API access to transCRISPR?
There is API access to status and results. After experiment start, the URL looks like https://transcrispr.igcz.poznan.pl/transcrispr/result/05eef993941b4452822f71737c9e45e0/The last part (05eef993941b4452822f71737c9e45e0) is unique hash value that can be used to check status and retrieve results.Results can be then checked using URL: https://transcrispr.igcz.poznan.pl/transcrispr/api-results/05eef993941b4452822f71737c9e45e0/?format=json and status with URL: https://transcrispr.igcz.poznan.pl/transcrispr/api-status/05eef993941b4452822f71737c9e45e0/?format=jsonStatus codes: WAITING = 0 STARTED = 1 PROCESSED_SEQUENCES = 5 PROCESSED_MOTIFS = 10 FOUND_MOTIFS = 20 OFF_TARGETS_02 = 25 OFF_TARGETS_03 = 27 OFF_TARGETS = 30 ON_TARGETS = 40 LOCALIZATION = 45 STATISTICS = 50 FINISHED = 80 ERROR = 500
Can I upload a custom genome?
Currently, it is not possible to upload a custom genome. Users may choose from 30 available genomes. If you need a genome which is not currently supported by transCRISPR but is available in UCSC, please send us a fasta file of the genome ordered by chromosomes. We will do our best but cannot guarantee that we will be able to add this genome to transCRISPR.
Can I design guides for sequence motif of interest for the whole genome?
It is not possible to mark the whole genome as target sequence in Step 3.
Is there a length limitation for sequence motif as input?
No. There is no required minimum or maximum length of sequence motif.
I used the “Display in the Genome Browser” button but only one guide is shown. What can I do?
TransCRISPR will automatically show you the first designed guide from the results. To see more guides in UCSC Genome Browser in the given region, try using “zoom out” button or simply paste the coordinates of interest.

To visualize all guides in the given region, be sure to mark “full” in display mode for the guides on- and off-target tracks.
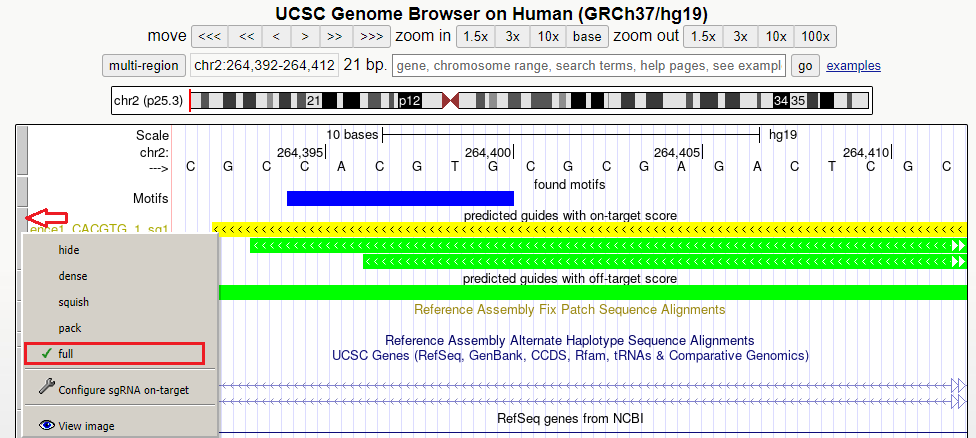
To navigate through guides on several positions and/or chromosomes use the start coordinate provided by transCRISPR in the results section “Found motifs and guides”.
How to cite transCRISPR?
Woźniak T, Sura W, Kazimierska M, Kasprzyk ME, Podralska M, Dzikiewicz-Krawczyk A.
TransCRISPR-sgRNA design tool for CRISPR/Cas9 experiments targeting specific sequence motifs. Nucleic Acids Res. 2023 May 9:gkad355. doi: 10.1093/nar/gkad355


